Understand human lung responses to infections
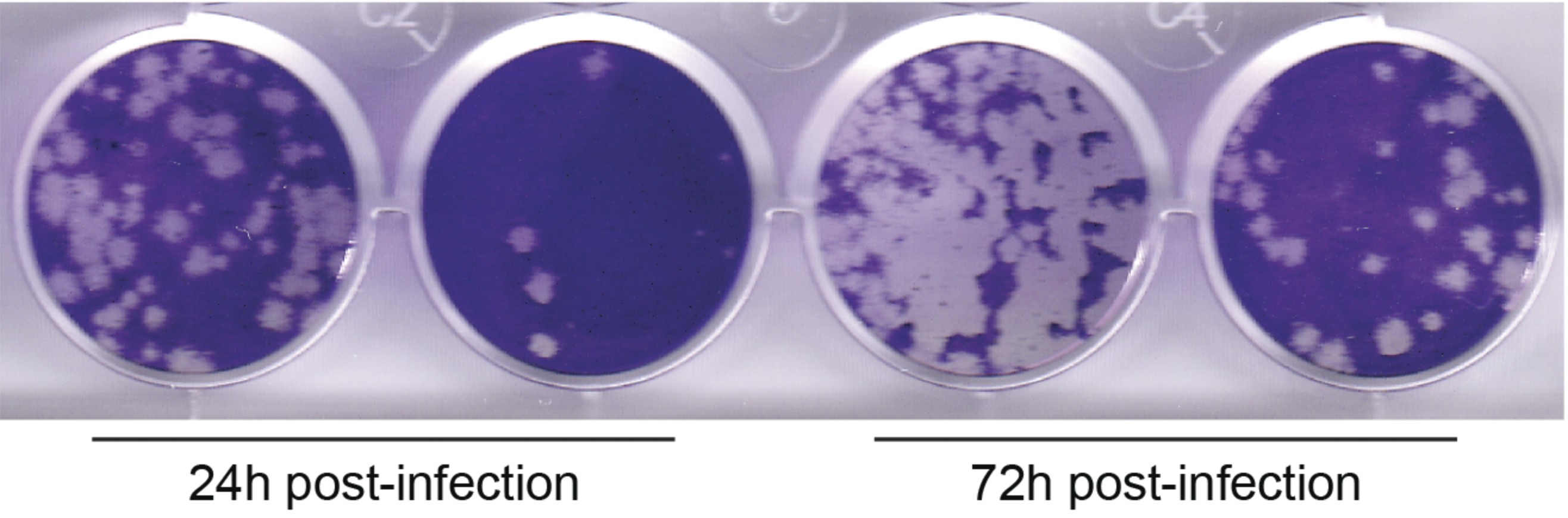
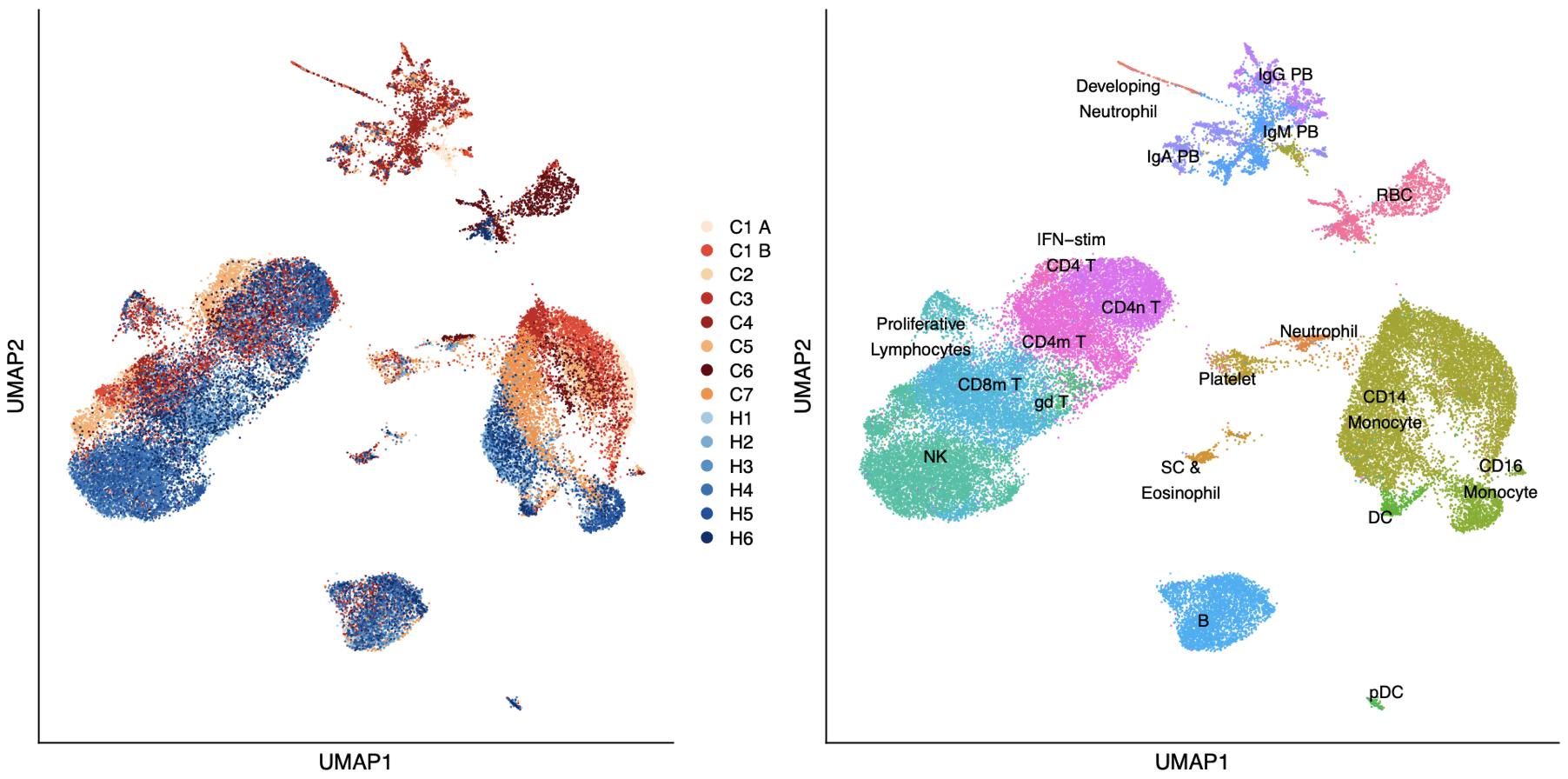
In the earliest stages of infection, we are interested in the interactions between pathogens, epithelium, and/or supporting immune cells. We use pathogens in the BSL2 and BSL3 with airway epithelial cell lines, human lung organoids, and explants to dissect lung pathogenesis, with the goal of highlighting targetable pathways. We have a special interest in human natural killer (NK) cells and macrophages. So far we have focused on SARS-CoV-2, including exploring novel entry pathways, but are expanding to other viral pathogens and fungal pathogens.